
 |
 |
|||
| A Brief Introduction to Antigenic Variation in African Trypanosomes | |||
| Antigenic variation is the primary cause of the persistence and lethality of African trypanosome infections (human Sleeping Sickness). A more detailed exposition of this subject will be forthcoming one day. |
Many viruses, bacteria and parasites have developed ways of evading our immune systems. Each year we are hit by new strains of influenza virus, which have mutated so they are no longer destroyed by the antibodies and T-cells that we developed to a previous infection. The African trypanosomes infect our blood, organs and tissues but, unlike viruses, they do not invade our cells. Therefore, they are continuously exposed to the antibodies that we generate against them, and which also circulate in our bloodstream. In fact, most of the circulating trypanosomes are successfully destroyed by these antibodies. The problem is that there are always survivors: trypanosomes that change their identity so they are not recognized by the current wave of antibodies. Unlike influenza or other viruses or bacteria, which need time to mutate to avoid destruction, trypanosomes have evolved a built-in system that quickly switches the antigens against which our immune system produces antibodies. This process is called Antigenic Variation, and has been most extensively investigated in Trypanosoma brucei, which is relatively easy to grow and study in the laboratory. Trypanosoma brucei is mainly a veterinary problem, but this species has two practically indistinguishable relatives, Trypanosoma rhodesiense and Trypanosoma gambiense, which cause virulent or chronic infections, respectively, in humans. |
||
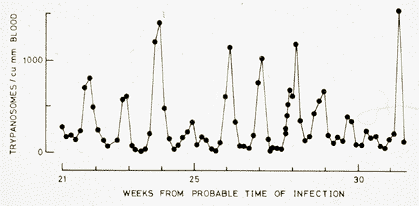 |
The graph to the left is redrawn from a report published in 1910, by Ross and Thomson, of the progress of a human infection with Trypanosoma gambiense. Despite repeated drug treatment, this patient eventually died, as do probably all humans and domestic animals infected with this virulent organism, in the absence of treatment. What the graph shows is that if one measures the number of trypanosomes circulating in the bloodstream (the parasitemia) at daily intervals, one detects a remarkable periodicity. This is due to antigenic variation. As the trypanosomes multiply (which they do by dividing into two daughter cells, about every 8 hours), we make antibodies against them. After 5 to 7 days, these antibodies destroy most of the trypanosomes, and the parasitemia decreases, but a new wave of trypanosomes appears. These are unaffected by antibodies generated against the previous wave, so the immnune system has to start over again. This process continues essentially indefinitely, until the death of the host. During this time, we continue to make primary responses against new antigens. One consequence of this is that our immune system becomes quite exhausted and our blood levels of antibody (immunoglobulin IgM) go sky high, to no useful effect. |
|
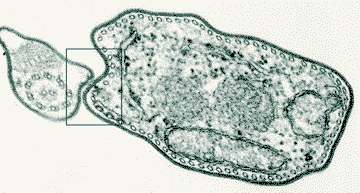
What are these antigens and how do they switch so easily? The first clue came from electron microscopy of trypanosomes. In 1979, Keith Vickerman drew attention to a feature of the surface of infective forms of Trypanosoma brucei. This was the presence of a Surface Coat, a regular dense structure that surrounded the entire cell body and flagellum of the trypanosome. Vickerman suggested that the Surface Coat might contain the antigens responsible for antigenic variation. The picture shows a thin slice through a trypanosome viewed in an electron microscope. The Surface Coat is represented by the fuzzy black outline of the cell in this photograph. It can be seen in more detail in a higher magnification of the boxed area. We discovered that the Surface Coat of every individual trypanosome is formed by 10 million copies of a single molecular species of antigen. These antigens are called glycoproteins, because they consist of a protein backbone with sugar (glyco) attachments. To Switch its Surface Coat, the trypanosome can select any member of a large family of what we call the Variant Surface Glycoprotein (VSG) family. It is the frequent switching of the VSG forming the Surface Coat that accounts for antigenic variation. Each VSG is encoded by an individual VSG Gene. Trypanosoma brucei contains hundreds of VSG Genes, accounting for about 10% of all its genes. If this system could be disrupted, our immune systems might be able to eliminate trypanosome infections. Much of our research is directed toward understanding how trypanosomes regulate antigenic variation. Trypanosomes appear to exert tight control over the expression of VSG genes. We don't know why this is so important to them, but it might be an intrinsic side effect of the switching mechanisms that the trypanosome has evolved. One of the central questions, therefore, is how does the trypanosome express only one of the hundreds of VSG genes at any time in any individual cell? Two important subsidiary questions are what regulates the timing of a switch and what regulates the order in which VSG genes are expressed? This kind of 'allelic choice' is not unique to trypanosomes. For example, detection of odors in the noses of animals involves the use of a large family of odorant receptors, but each sensory cell in the nose expresses only one member of this family, despite containing all of the genes. There are other examples, in animals, of cells that are able to selectively express single genes from families of various complexity. In trypanosomes, the VSG gene that is expressed is always located very close to the end of a chromosome (a telomere). This is a necessary but not sufficient condition to be expressed because about 20 telomeres contain VSG genes in the necessary configuration to be expressed (trypanosomes have two copies of at least 11 chromosomes, hence they have at least 44 telomeres). In some respects, this location requirement reduces the complexity of the exclusiveness question to why is only one out of 20 telomeric VSG genes expressed. We know there are two main mechanisms by which a VSG gene can be switched. One can switch the telomeric Expression Site that is being transcribed or one can shuttle VSG genes, from other locations in the chromosomes, into an active Expression Site. More technical details of these mechanisms can be found elsewhere on this site. |
 |
|
| Space is reserved for a VSG picture. | ||